Diagnóstico citogenético de AF en una cohorte de pacientes con características clínicas de sospecha de anemia de Fanconi
Contenido principal del artículo
Resumen
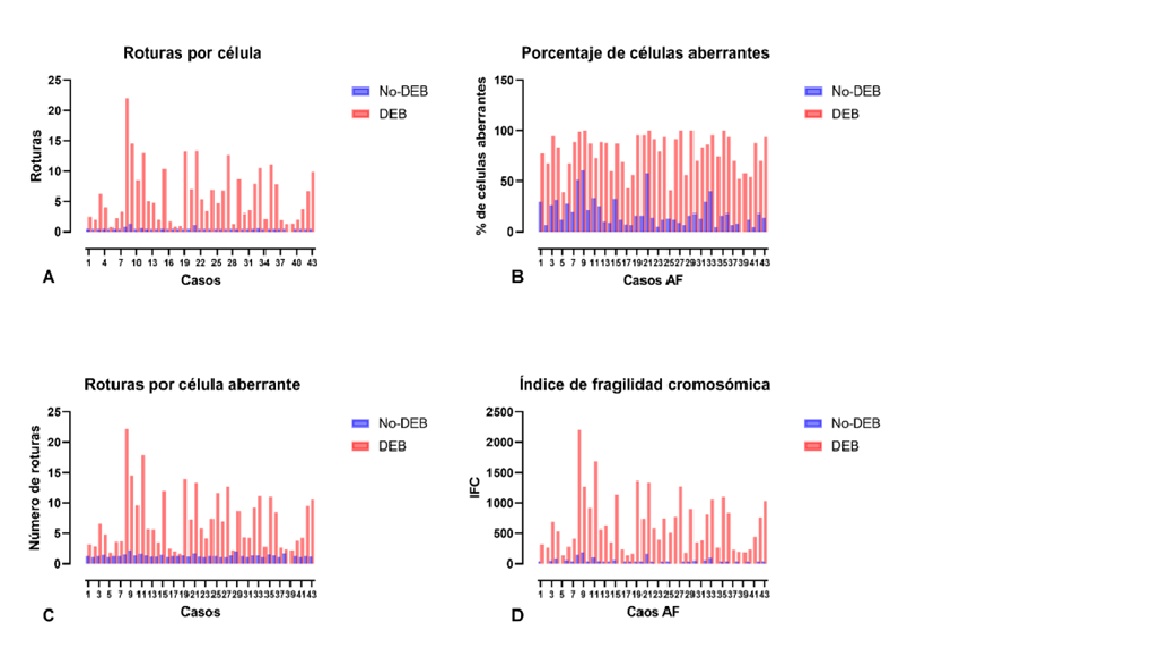
Antecedentes: la Anemia de Fanconi (AF) es una enfermedad heredada, que afecta la reparación del ADN. Clínicamente es heterogénea; mayoritariamente se presentan malformaciones congénitas, aplasia medular temprana y predisposición a cáncer. El defecto genético causa hipersensibilidad a genotóxicos e inestabilidad cromosómica. Esta característica se considera el mejor marcador diagnóstico; sin embargo, llegar a él puede convertirse en un desafío. Objetivo: caracterizar pacientes con AF mediante pruebas citogenéticas en individuos con rasgos clínicos sugestivos de la enfermedad. Métodos: se analizaron 157 individuos con sospecha clínica de AF, 19 con asociación VACTERL, 15 hermanos, y 34 individuos sanos. Se realizó registro de datos clínicos, y prueba citogenética con Diepoxibutano (DEB). Resultados: se identificaron 43 afectados por AF. La relación de índices en células tratadas con DEB del grupo AF vs. No-AF fue significativamente incrementada, 6.7 veces la proporción de células aberrantes, 48 veces el número de roturas por célula, y 6.3 veces el número de roturas por célula aberrante. En AF la edad media de muestreo fue 9.2 años, la proporción de sexos M:F 1.5:1, consanguinidad en 11 casos. Los sistemas hematológico, esquelético, tegumentario, y urinario estuvieron significativamente alterados. Conclusiones: La AF se identificó en 26 % del grupo de sospecha y en 13 % de hermanos sin sospecha previa. La enfermedad hematológica fue el síntoma más recurrente presente en 93 % de los casos, y fue principalmente la primera sospecha de AF y motivo de estudio genético.
Descargas
Detalles del artículo

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Licencia Creative Commons
Atribución-NoComercial-CompartirIgual 4.0 Internacional (CC BY-NC-SA 4.0)
Usted es libre de:
Compartir - copiar y redistribuir el material en cualquier medio o formato.
Adaptar - remezclar, transformar y construir a partir del material
La licencia no puede revocar estas libertades en tanto se sigan los términos de la licencia.
- Atribución — Usted debe dar crédito de manera adecuada, brindar un enlace a la licencia, e indicar si se han efectuado cambios. Puede hacerlo en cualquier forma razonable, pero no de forma tal que sugiera que usted o su uso tienen el apoyo del licenciante.
- NoComercial — Usted no puede hacer uso del material con propósitos comerciales.
- CompartirIgual— Si remezcla, transforma o crea a partir del material, debe distribuir su contribución bajo la misma licencia del original.
- No hay restricciones adicionales — No puede aplicar términos legales ni medidas tecnológicas que restrinjan legalmente a otras a hacer cualquier uso permitido por la licencia.
Citas
Fanconi Anemia - GeneReviews® - NCBI Bookshelf [Internet]. https://www.ncbi.nlm.nih.gov/books/NBK1401/ [citado 2022 Jul 27]
Sumpter R, Levine B. Novel functions of Fanconi anemia proteins in selective autophagy and inflammation. Oncotarget 2016; 7(32):50820–1. DOI: https://doi.org/10.18632/oncotarget.10970
Milletti G, Strocchio L, Pagliara D, Girardi K, Carta R, Mastronuzzi A, et al. Canonical and noncanonical roles of fanconi anemia proteins: Implications in cancer predisposition. Cancers (Basel) 2020; 12(9):2684 DOI: https://doi.org/10.3390/cancers12092684
Rosenberg PS, Tamary H, Alter BP. How High are Carrier Frequencies of Rare Recessive Syndromes? Contemporary Estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A 2012; 155(8):1877–83 DOI: https://doi.org/10.1002/ajmg.a.34087
Callén E, E, Casado JA, Tischkowitz MD, Bueren JA, Creus A, Marcos R, et al. A common founder mutation in FANCA underlies the world’s highest prevalence of Fanconi anemia in Gypsy families from Spain. Blood 2005; 105(5):1946–9 DOI: https://doi.org/10.1182/blood-2004-07-2588
Kutler D, Auerbach A. Fanconi anemia in Ashkenazi Jews. Fam Cancer 2004; 3(3–4):241–8. DOI: https://doi.org/10.1007/s10689-004-9565-8
Morgan N V., Essop F, Demuth I, De Ravel T, Jansen S, Tischkowitz M, et al. A common Fanconi anemia mutation in black populations of sub-Saharan Africa. Blood 2005; 105(9):3542–4 DOI: https://doi.org/10.1182/blood-2004-10-3968
Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res 2009; 668(1–2):4–10 DOI: https://doi.org/10.1016/j.mrfmmm.2009.01.013
Mikat B, Roll C, Schindler D, Gembruch U, Klempert I, Buiting K, et al. X-linked recessive VACTERL-H due to a mutation in FANCB in a preterm boy. Clin Dysmorphol 2016; 25(2):73–6 DOI: https://doi.org/10.1097/MCD.0000000000000111
Giampietro PF, Adler-Brecher B, Verlander PC, Pavlakis SG, Davis JG, Auerbach AD. The need for more accurate and timely diagnosis in Fanconi anemia: A report from the International Fanconi Anemia Registry. Pediatrics. 1993; 91(6):1116–20 DOI: https://doi.org/10.1542/peds.91.6.1116
Wegman-Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol 2017; 177(4):526–42 DOI: https://doi.org/10.1111/bjh.14535
Alter BP, Rosenberg PS. VACTERL-H association and fanconi anemia. Mol Syndromol 2013; 4(1–2):87–93 DOI: https://doi.org/10.1159/000346035
Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA inter strand cross-link repair. Cold Spring Harb Perspect Biol 2013; 3(10):a012732 DOI: https://doi.org/10.1101/cshperspect.a012732
Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr Opin Cell Biol 2015; 37:49–60 DOI: https://doi.org/10.1016/j.ceb.2015.09.002
García-De-teresa B, Rodríguez A, Frias S. Chromosome instability in fanconi anemia: From breaks to phenotypic consequences. Genes (Basel) 2020; 11(12):1–35 DOI: https://doi.org/10.3390/genes11121528
Bogliolo M, Surrallés J. Fanconi anemia: A model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev 2015; 33:32–40 DOI: https://doi.org/10.1016/j.gde.2015.07.002
Che R, Zhang J, Nepal M, Han B, Fei P. Multifaceted Fanconi Anemia Signaling. Trends Genet 2018; 34(3):171–83 DOI: https://doi.org/10.1016/j.tig.2017.11.006
Datta A, Brosh RM. Holding all the cards—how fanconi anemia proteins deal with replication stress and preserve genomic stability. Genes (Basel) 2019; 10(2):170. DOI: https://doi.org/10.3390/genes10020170
Moreno OM, Paredes AC, Suarez-Obando F, Rojas A. An update on Fanconi anemia: Clinical, cytogenetic and molecular approaches (review). Biomed Reports 2021; 15(3):1–10 DOI: https://doi.org/10.3892/br.2021.1450
Nepal M, Che R, Zhang J, Ma C, Fei P. Fanconi Anemia Signaling and Cancer. Trends Cancer 2017; 3(12):840-856 DOI: https://doi.org/10.1016/j.trecan.2017.10.005
Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet 2004; 36(11):1219–24 DOI: https://doi.org/10.1038/ng1458
Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat Rev Mol Cell Biol 2016; 17(6):337–49 DOI: https://doi.org/10.1038/nrm.2016.48
Peake JD, Noguchi E. Fanconi anemia: current insights regarding epidemiology, cancer, and DNA repair. Hum Genet 2022 May 21. doi: 10.1007/s00439-022-02462-9. Epub ahead of print. DOI: https://doi.org/10.1007/s00439-022-02462-9
Bogliolo M, Pujol R, Aza-Carmona M, Muñoz-Subirana N, Rodriguez-Santiago B, Casado JA, et al. Optimised molecular genetic diagnostics of Fanconi anaemia by whole exome sequencing and functional studies. J Med Genet 2020; 57(4):258-268 DOI: https://doi.org/10.1136/jmedgenet-2019-106249
Auerbach AD, Rogatko A, Schroeder-Kurth TM. International Fanconi Anemia Registry: Relation of clinical symptoms to diepoxybutane sensitivity. Blood 1989; 73(2):391–6 DOI: https://doi.org/10.1182/blood.V73.2.391.bloodjournal732391
Krausz C, Riera-Escamilla A, Chianese C, Moreno-Mendoza D, Ars E, Rajmil O, et al. From exome analysis in idiopathic azoospermia to the identification of a high-risk subgroup for occult Fanconi anemia. Genet Med 2019; 21(1):189–94 DOI: https://doi.org/10.1038/s41436-018-0037-1
Castella M, Pujol R, Callén E, Ramírez MJ, Casado JA, Talavera M, et al. Chromosome fragility in patients with Fanconi anaemia: Diagnostic implications and clinical impact. J Med Genet 2011; 48(4):242–50 DOI: https://doi.org/10.1136/jmg.2010.084210
Moreno OM, Sánchez AI, Herreño A, Giraldo G, Suárez F, Prieto JC, et al. Phenotypic Characteristics and Copy Number Variants in a Cohort of Colombian Patients with VACTERL Association. Mol Syndromol 2020; 11:271–83 DOI: https://doi.org/10.1159/000510910
Auerbach AD. Diagnosis of Fanconi Anemia by diepoxybutane analysis. Curr Protoc Hum Genet 2015; 2015:8.7.1-8.7.17. DOI: https://doi.org/10.1002/0471142905.hg0807s85
Swift MR and Hirschhron K. FA’s Inherited Susceptibility to chromosome breakage. Ann Intern Med 1966; 65(3):496-503 DOI: https://doi.org/10.7326/0003-4819-65-3-496
Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature 1976; 261(5560):494–6 DOI: https://doi.org/10.1038/261494a0
Aslan D. Failure or delay in diagnosing Fanconi anemia - A well-defined genetic disorder. Turk J Pediatr 2013; 55(4):462–4
Oostra AB, Nieuwint AWM, Joenje H, De Winter JP. Diagnosis of fanconi anemia: Chromosomal breakage analysis. Anemia 2012; 2012:238731 DOI: https://doi.org/10.1155/2012/238731
Esmer C, Sánchez S, Ramos S, Molina B, Frias S, Carnevale A. DEB Test for Fanconi Anemia Detection in Patients with Atypical Phenotypes. Am J Med Genet 2004; 124A(1):35–9 DOI: https://doi.org/10.1002/ajmg.a.20327
Reutter H, Ludwig M. VATER/VACTERL association: Evidence for the role of genetic factors. Mol Syndromol 2013; 4(1–2):16–9 DOI: https://doi.org/10.1159/000345300
Trujillo JP, Surralles J. Savior siblings and Fanconi anemia: analysis of success rates from the family’s perspective. Genet Med 2015; 17:935–8 DOI: https://doi.org/10.1038/gim.2014.206
Korgaonkar S, Ghosh K, Jijina F, Vundinti BR. Chromosomal breakage study in children suspected with fanconi anemia in the indian population. J Pediatr Hematol Oncol 2010; 32(8):606–10 DOI: https://doi.org/10.1097/MPH.0b013e3181e8865f
Cirkovic S, Guc-Scekic M, Vujic D, Ilic N, Micic D, Skoric D, et al. Diagnosis of Fanconi’s anemia by diepoxybutane analysis in children from Serbia. Balk J Med Genet 2011; 14(2):65–70 DOI: https://doi.org/10.2478/v10034-011-0048-6
Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev 2010; 24(3):101–22 DOI: https://doi.org/10.1016/j.blre.2010.03.002
Zierhut HA, Tryon R, Sanborn EM. Genetic Counseling for Fanconi Anemia: Crosslinking Disciplines. J Genet Couns 2014; 23(6):910–21 DOI: https://doi.org/10.1007/s10897-014-9754-z
Auerbach AD. A test for Fanconi’s anemia. Blood 1988; 72(1):366–9. DOI: https://doi.org/10.1182/blood.V72.1.366.bloodjournal721366
Fargo JH, Rochowski A, Giri N, Savage SA, Olson SB, Alter BP. Comparison of chromosome breakage in non-mosaic and mosaic patients with Fanconi anemia, relatives, and patients with other inherited bone marrow failure syndromes. Cytogenet Genome Res 2014; 144(1):15–27 DOI: https://doi.org/10.1159/000366251
Nicoletti E, Rao G, Bueren JA, Río P, Navarro S, Surrallés J, et al. Mosaicism in Fanconi anemia: concise review and evaluation of published cases with focus on clinical course of blood count normalization. Ann Hematol 2020; 99:913–24 DOI: https://doi.org/10.1007/s00277-020-03954-2
Revy P, Kannengiesser C, Fischer A. Somatic genetic rescue in Mendelian haematopoietic diseases. Nat Rev Genet 2019; 20(10):582–98 DOI: https://doi.org/10.1038/s41576-019-0139-x
Waisfisz Q, Morgan N V., Savino M, De Winter JP, Van Berkel CGM, Hoatlin ME, et al. Spontaneous functional correction of homozygous Fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet 1999; 22(4):379–83 DOI: https://doi.org/10.1038/11956
Soulier J, Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood 2005 ; 105(3):1329–36 DOI: https://doi.org/10.1182/blood-2004-05-1852
Mankad A, Taniguchi T, Cox B, Akkari Y, Rathbun RK, Lucas L, et al. Natural gene therapy in monozygotic twins with Fanconi anemia. Blood 2006; 107(8):3084–90 DOI: https://doi.org/10.1182/blood-2005-07-2638
Ramírez MJ, Pujol R, Trujillo-Quintero JP, Minguillon J, Bogliolo M, Rio P, et al. Natural gene therapy by reverse mosaicism leads to improved hematology in Fanconi anemia patients. Am J Hematol 2021; 96:989–999 DOI: https://doi.org/10.1002/ajh.26234
Hyde RK, Liu PP. GATA2 mutations lead to MDS and AML. Nat Genet 2011; 43(10):926–7 DOI: https://doi.org/10.1038/ng.949
Sarkar K, Han SS, Wen KK, Ochs HD, Dupré L, Seidman MM, et al. R-loops cause genomic instability in T helper lymphocytes from patients with Wiskott-Aldrich syndrome. J Allergy Clin Immunol 2018; 142(1):219–34 DOI: https://doi.org/10.1016/j.jaci.2017.11.023
Mehta PA, Harris RE, Davies SM, Kim MO, Mueller R, Lampkin B, et al. Numerical chromosomal changes and risk of development of myelodysplastic syndrome-acute myeloid leukemia in patients with Fanconi anemia. Cancer Genet Cytogenet 2010; 203(2):180–6 DOI: https://doi.org/10.1016/j.cancergencyto.2010.07.127
Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. Hematologic abnormalities in Fanconi anemia: An International Fanconi Anemia Registry study. Blood 1994; 84(5):1650–5 DOI: https://doi.org/10.1182/blood.V84.5.1650.bloodjournal8451650
Cioc Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: Morphologic and cytogenetic characteristics. Am J Clin Pathol 2010; 133(1):92–100 DOI: https://doi.org/10.1309/AJCP7W9VMJENZOVG
Rochowski A, Olson SB, Alonzo TA, Gerbing RB, Lange BJ, Alter BP. Patients with Fanconi anemia and AML have different cytogenetic clones than de novo cases of AML. Pediatr Blood Cancer 2012; 59(5):922–4 DOI: https://doi.org/10.1002/pbc.24168
Taylor AMR, Rothblum-Oviatt C, Ellis NA, Hickson ID, Meyer S, Crawford TO, et al. Chromosome instability syndromes. Nat Rev Dis Primers 2019; 19;5(1):64 DOI: https://doi.org/10.1038/s41572-019-0113-0
Vásquez Palacio G, Ramírez Castro JL, Posada Díaz Á, Sierra M, Botero OL, Durango NE, et al. Leucemia linfoide aguda: Estudio citogenético en niños atendidos en el Hospital Universitario San Vicente de Paúl de Medellín en el período 1998-2001. Iatreia 2002; 15(4):217–25 DOI: https://doi.org/10.17533/udea.iatreia.3962
Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 2003; 101(4):1249–56 DOI: https://doi.org/10.1182/blood-2002-07-2170
Fiesco-roa MO, Giri N, Mcreynolds LJ, Best AF, Alter BP. Blood Reviews Genotype-phenotype associations in Fanconi anemia : A literature review. Blood Rev 2019; 37:100589 DOI: https://doi.org/10.1016/j.blre.2019.100589
Smith AR, Wagner JE. Current clinical management of Fanconi anemia. Expert Rev Hematol. 2012;5(5):513–22. DOI: https://doi.org/10.1586/ehm.12.48
Giri N, Batista DL, Batista DL, Alter BP, Stratakis CA. Endocrine Abnormalities in Patients with Fanconi Anemia. J Clin Endocrinol Metab 2007; 92(7):2624-31 DOI: https://doi.org/10.1210/jc.2007-0135
Huck Hanenberg H, Gudowius S, Fenk R, Kalb R, Neveling K, et al. Delayed diagnosis and complications of Fanconi anaemia at advanced age - A paradigm. Br J Haematol 2006; 133(2):188–97 DOI: https://doi.org/10.1111/j.1365-2141.2006.05998.x